This directory contains supplementary material and data for the paper:
S. Mahony, P.E. Auron, P.V. Benos, "DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies", PLoS Comput Biol (2007) 3:e61.
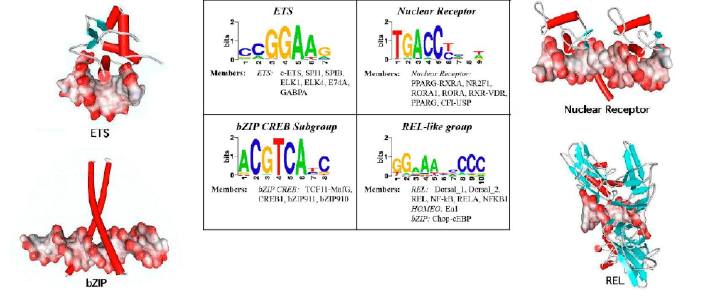
The supplementary material included here is the table with the complete results of the comparison of 5 different column-to-column distance metrics, 2 PSSM model alignment strategies and various gap penalties. The data deposited here include the set of JASPAR PSSM models we used for the testings and the list of the TRANSFAC transcription factors. For each transcription factor we have included information related to the structural family it belongs.
A copy of the paper can be found here
For software download, please click here
- Supplementary Table
Performance of various aligning strategies and column-to-column comparison metrics.
- JASPAR matrices
JASPAR (version 2005) matrices for all 96 transcription factors we used in the paper. The matrices are provided in the TRANSFAC format. Each DE line contains the TF name, TF structural group, species and taxonomy information.
- JASPAR non-zinc finger matrices
JASPAR (version 2005) matrices for the 71 non-zinc finger transcription factors. The matrices are provided in the TRANSFAC format. Each DE line contains the TF name, TF structural group, species and taxonomy information.
- List of TRANSAFC matrices
List of the TRANSFAC (version 9.3) matrices we used in the paper. The list contains the TF name, TF structural group (see TF_Class_Lookup.txt file for correspondence), and the TRANSFAC accession number.
- List of non-zinc finger TRANSAFC matrices
Same as above without the zinc finger TFs.
- TRANSFAC class lookup table
Lookup table for the TRANSFAC structural group codes.
- Random matrices
Randomly generated PSSM models from JASPAR columns (10,000 matrices in TRANSFAC format)
|

