This directory contains supplementary material and data for the paper:
S. Kadri, V. Hinman, P.V. Benos, "HHMMiR: Efficient de novo Prediction of MicroRNAs using Hierarchical Hidden Markov Models", BMC Bioinformatics (Proc APBC 2009) (2009) 10 (Suppl 1):S35.
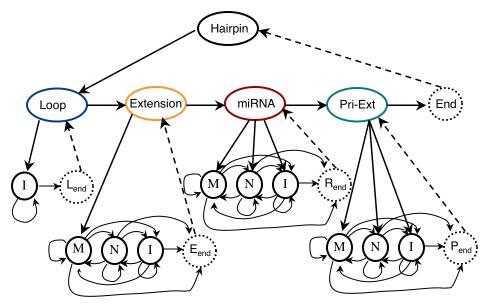
The supplementary material included here is summary characteristics of regions of miRNA stemloops for various species, detailed description of the HHMM model, and the training and testing datasets used in this study.
A copy of the paper can be found here
For software download, please click here
|

