This directory contains supplementary material and data for the paper:
D.L. Corcoran, E. Feingold, J. Dominick, M. Wright, J. Harnaha, M. Trucco, N. Giannoukakis and P.V. Benos, "Footer: a quantitative comparative genomics method for efficient recognition of cis-regulatory elements", Genome Res (2005) 15:840-847.
The supplementary material included here are tables with the complete list of predictions of FOOTER and results from detailed comparison with other methods. The data deposited here include the set of promoters the analysis was based (human and mouse or rat), the corresponding DBA alignments and the PSSM models we used for this analysis.
A copy of the paper can be found here
- Supplementary Table 1 (version 2.1, June 16th, 2005):
Complete list of predictions of FOOTER on 72 known binding sites of 19 transcription factors in 24 promoters. The known sites and their positions in the promoters are reported. Version 2.1 also contains the species where the reported sequence comes from and the reference list that was omitted by mistake in the the original version.
- Supplementary Table 2 (version 2, June 11th, 2005):
Comparison of FOOTER with other methods on the above test set.
- Promoter regions (test set)
The 14 promoter regions analysed.
- DBA alignments
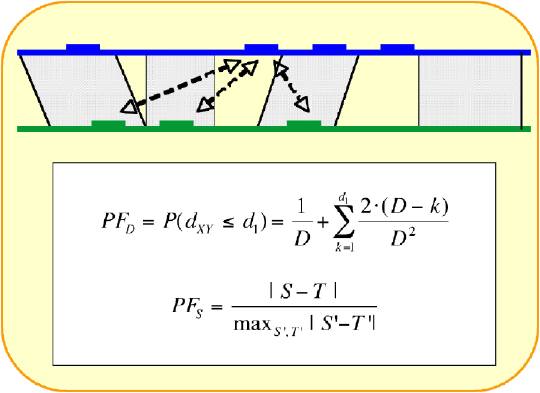
The DBA alignments of the 14 human-rodent promoter region comparisons. FOOTER uses the DBA alignments to calculate the distance between two putative binding sites.
- PSSM models
The PSSM models used in our analysis. These models are provided with permission from BIOBASE.
|

